Overview of Japanese Encephalitis Virus Structure
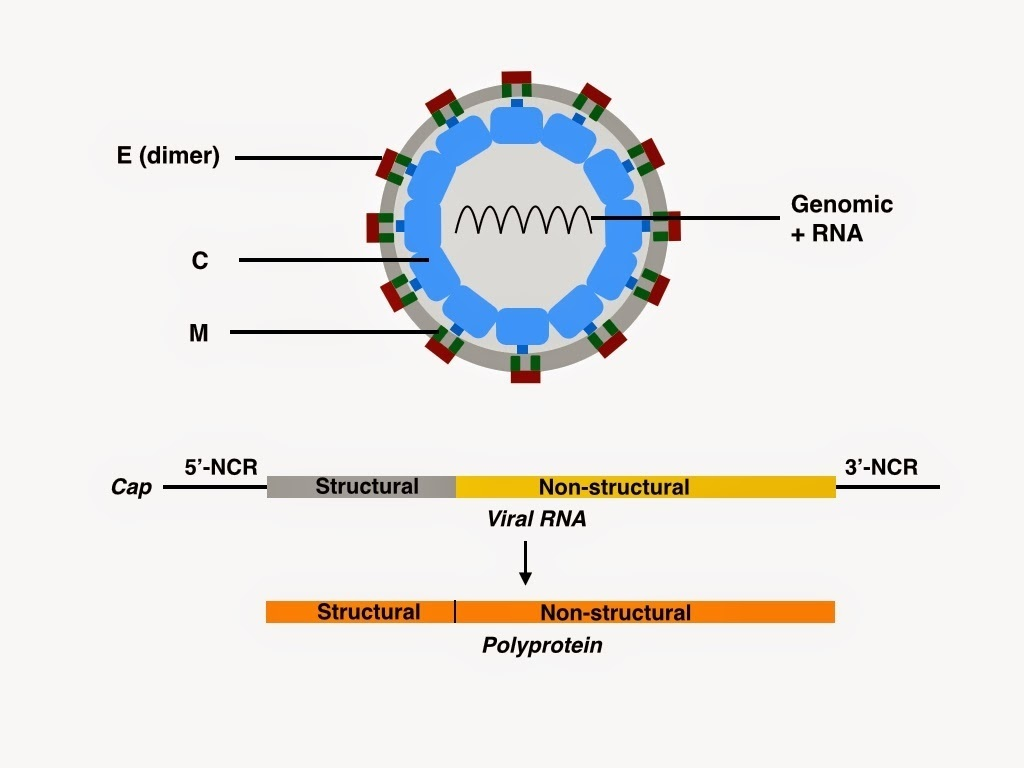
The Japanese encephalitis virus (JEV) is a positive-sense, single-stranded RNA virus (~11 kb genome) belonging to the Flaviviridae family, with an enveloped icosahedral structure.
Genome Organization of Japanese Encephalitis Virus
The genome encodes a single polyprotein (capsid C, precursor membrane prM/M, and envelope E) and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5).
Key Structural Features of Japanese Encephalitis Virus
Its key structural features are
1) Protein Capsid
The virion is roughly spherical, about 50 mm in diameter, with an electron-dense nucleocapsid core (~30 nm diameter) surrounded by a lipid bilayer derived from the host cell membrane.
2) Genome
The genome is a single-stranded positive-sense RNA (~11kb ) enclosed within the nucleocapsid, encoding three structural proteins and seven nonstructural proteins.
3) Three Structural Proteins
The three structural proteins are
Capsid (C) Protein
Forms the nucleocapsid by encapsulating the RNA genome. It exists as a homodimer with a helical structure and is critical for nucleic acid binding and virus assembly.
Envelope (E) Protein
The major surface glycoprotein is responsible for receptor binding, membrane fusion, and induction of neutralization antibodies. It is organized into three domains, including a fusion loop for membrane fusion during cell entry.
Membrane (M) Protein
Present as the precursor prM in immature virions, it acts as a chaperone preventing fusion. Cleavage of prM to M during maturation exposes the fusion loop of the E protein, allowing infectivity.
Viral Envelope and Entry Mechanism
The viral envelope is studded with E and M proteins embedded in the lipid bilayer that surrounds the nucleocapsid. The viral entry involves attachment via the E protein to host cell receptors, internalization by endocytosis, fusion of the virus membrane with host endosomal membranes, and release of the nucleocapsid into the cytoplasm. Japanese Encephalitis Virus